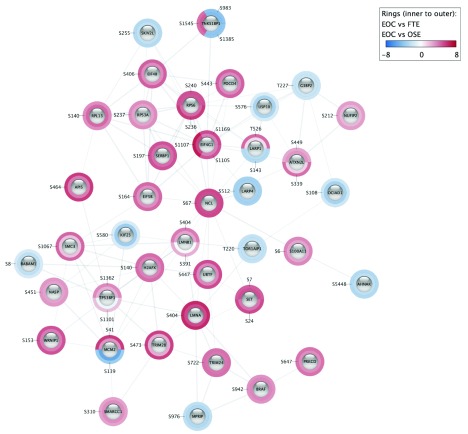
Cytoscape is an open-source software program used to research and visualize organic networks. Along with having the ability to import networks from quite a lot of sources, Cytoscape permits customers to import tabular node information and visualize it onto networks.
Sadly, such information tables can solely include one row of information per node, whereas omics information typically have a number of rows for a similar gene or protein, representing completely different post-translational modification websites, peptides, splice isoforms, or situations.
Right here, we current a brand new app, Omics Visualizer, that permits customers to import information tables with a number of rows referring to the identical node, join them to a number of networks, and visualize the linked information onto networks.
Omics Visualizer makes use of the Cytoscape enhancedGraphics app to point out the information both within the nodes (pie visualization) or across the nodes (donut visualization), the place the colours of the slices characterize the imported values. If the person doesn’t present a community, the app can retrieve one from the STRING database utilizing the Cytoscape stringApp.
Visualize omics data on networks with Omics Visualizer, a Cytoscape App.
Fragrant stacking facilitated self-assembly of ultra-short ionic complementary peptide sequence: β-sheet nanofibres with outstanding gelation and interfacial properties.
Understanding peptide self-assembly mechanisms and stability of the fashioned assemblies is essential for improvement of useful nanomaterials.
Herein, we have now adopted rational design strategy to exhibit how minimal structural modification to a non-assembling ultra-short ionic self-complementary tetrapeptide FEFOkay (Phe4) remarkably enhanced stability of self-assembly into β-sheet nanofibres and induced hydrogelation.
This was achieved by changing versatile phenylalanine residue (F) by the inflexible phenylglycine (Phg) leading to constrained analogue PhgEPhgOkay (Phg4), which positioned fragrant rings in an orientation beneficial for fragrant stacking. Phg4 self-assembly into steady β-sheet ladders was facilitated by π-staking of fragrant sidechains alongside hydrogen bonding between spine amides alongside the nanofibre axis.
The contribution of those non-covalent interactions in stabilising self-assembly was predicted by in silico modelling utilizing molecular dynamics simulations and semi-empirical quantum mechanics calculations.
In aqueous medium, Phg4 β-sheet nanofibres entangled at a essential gelation focus > 20 mg/mL forming a community of nanofibrous hydrogel. Phg4 additionally demonstrated distinctive floor exercise in presence of immiscible oils and was superior to industrial emulsifiers in stabilising oil-in-water emulsions.
This was attributed to interfacial adsorption of amphiphilic nanofibrilles forming nanofibrillised microspheres. To our information, Phg4 is the shortest ionic self-complementary peptide rationally designed to self-assemble into steady β-sheet nanofibres able to gelation and emulsification.
Our outcomes recommend that Extremely-short Ionic-complementary Constrained Peptides or UICPs have important potential for the event of cost-effective, sustainable and multifunctional delicate bionanomaterials.